Radon example from Gelman and Hill (2006)
Contents
Radon example from Gelman and Hill (2006)¶
This is a reworking of the radon example from pymc3 https://docs.pymc.io/notebooks/multilevel_modeling.html
Other implementations in:
tensorflow https://www.tensorflow.org/probability/examples/Multilevel_Modeling_Primer - pymc3 https://docs.pymc.io/notebooks/multilevel_modeling.html
stan https://mc-stan.org/users/documentation/case-studies/radon.html
pyro https://github.com/pyro-ppl/pyro-models/blob/master/pyro_models/arm/radon.py
numpyro fibrosis dataset http://num.pyro.ai/en/stable/tutorials/bayesian_hierarchical_linear_regression.html
import altair as alt
try:
alt.renderers.enable('altair_saver', fmts=['png'])
except Exception:
pass
from bayes_window import BayesWindow, BayesConditions, LMERegression, BayesRegression
from utils import load_radon
df = load_radon()
df
| county | radon | floor | |
|---|---|---|---|
| 0 | AITKIN | 2.2 | 1 |
| 1 | AITKIN | 2.2 | 0 |
| 2 | AITKIN | 2.9 | 0 |
| 3 | AITKIN | 1.0 | 0 |
| 4 | ANOKA | 3.1 | 0 |
| ... | ... | ... | ... |
| 922 | WRIGHT | 6.4 | 0 |
| 923 | WRIGHT | 4.5 | 0 |
| 924 | WRIGHT | 5.0 | 0 |
| 925 | YELLOW MEDICINE | 3.7 | 0 |
| 926 | YELLOW MEDICINE | 2.9 | 0 |
919 rows × 3 columns
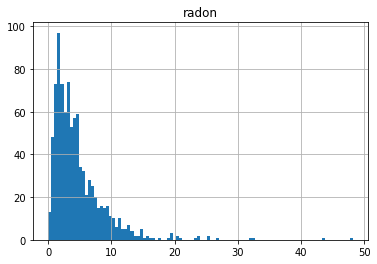
df.set_index(['county','floor']).hist(bins=100);
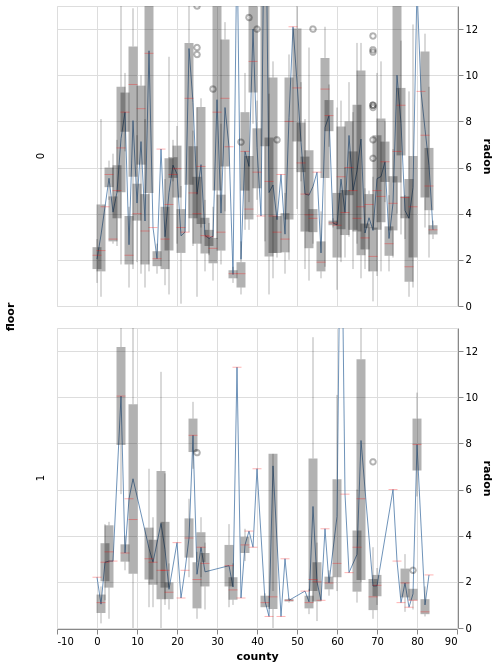
window=BayesWindow(df.reset_index(), y='radon', treatment='floor',group='county')
Plot data¶
window.plot(x='county').facet(row='floor')
Fit LME¶
lme=LMERegression(window)#formula='radon ~ floor + ( 1 | county)')
window.plot()
Fit Bayesian hierarchical with and without county-specific intercept¶
window1=BayesRegression(df=df.reset_index(), y='radon', treatment='floor',group='county')
window1.fit(add_group_intercept=True);
window1.plot()
2021-11-30 15:37:12.522319: E external/org_tensorflow/tensorflow/stream_executor/cuda/cuda_driver.cc:271] failed call to cuInit: CUDA_ERROR_COMPAT_NOT_SUPPORTED_ON_DEVICE: forward compatibility was attempted on non supported HW
Uneven number of entries in conditions! This will lead to nans in data (window.data["radon diff"(170, 120)
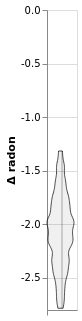
window1.plot(x=':O')
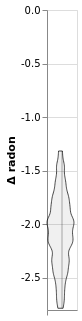
Inspect intercepts (horizontal ticks)¶
window1.plot_intercepts()

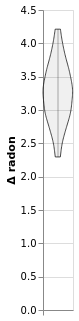
window2=BayesRegression(df=df.reset_index(), y='radon', treatment='floor',group='county')
window2.fit(add_group_intercept=False, add_group_slope=False, do_make_change='subtract');
window2.plot()
Uneven number of entries in conditions! This will lead to nans in data (window.data["radon diff"(170, 120)
(window.plot().properties(title='LME')|
window1.plot().properties(title='Partially pooled Bayesian')|
window2.plot().properties(title='Unpooled Bayesian'))
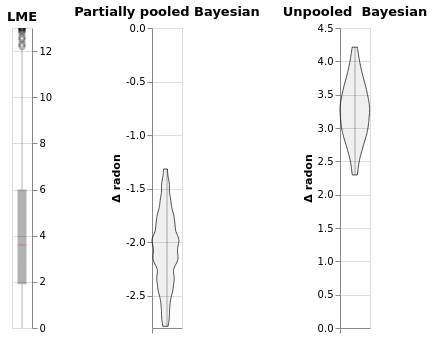
Compare the two models¶
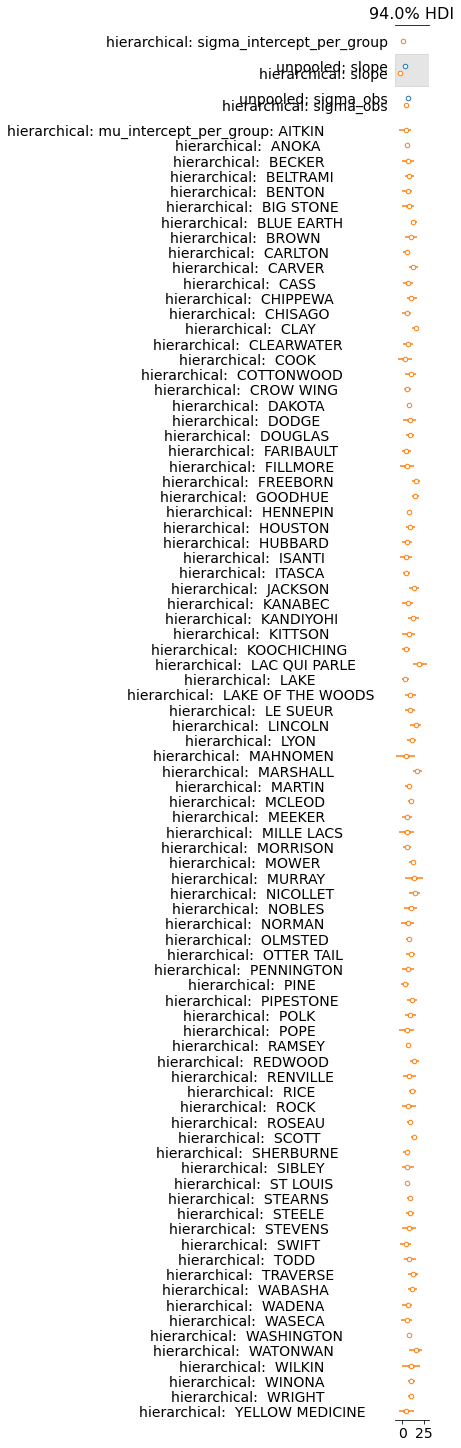
import arviz as az
datasets = {'unpooled' : window2.trace.posterior,
'hierarchical': window1.trace.posterior}
az.plot_forest(data=list(datasets.values()), model_names=list(datasets.keys()),
#backend='bokeh',
#kind='ridgeplot',
#ridgeplot_overlap=1.6,
combined=True);
For leave-one-out, let’s remove any counties that did not contain both floors. This drops about 250 rows
import pandas as pd
df_clean = pd.concat([ddf for i, ddf in df.groupby(['county'])
if (ddf.floor.unique().size>1)
and (ddf[ddf['floor']==0].shape[0]>1)
and (ddf[ddf['floor']==1].shape[0]>1)
])
df_clean
| county | radon | floor | |
|---|---|---|---|
| 4 | ANOKA | 3.1 | 0 |
| 5 | ANOKA | 2.5 | 0 |
| 6 | ANOKA | 1.5 | 0 |
| 7 | ANOKA | 1.0 | 0 |
| 8 | ANOKA | 0.7 | 0 |
| ... | ... | ... | ... |
| 907 | WINONA | 10.2 | 0 |
| 908 | WINONA | 11.3 | 0 |
| 909 | WINONA | 7.6 | 0 |
| 910 | WINONA | 11.8 | 0 |
| 911 | WINONA | 0.5 | 1 |
660 rows × 3 columns
window1.data=df_clean
window1.explore_models()
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
/tmp/ipykernel_2829554/1390140151.py in <module>
1 window1.data=df_clean
----> 2 window1.explore_models()
~/mmy/bayes-window/bayes_window/slopes.py in explore_models(self, parallel, add_group_slope, **kwargs)
410 'group': self.window.group,
411 'add_group_slope': True}])
--> 412 return compare_models(
413 df=self.window.data,
414 models=models,
~/mmy/bayes-window/bayes_window/model_comparison.py in compare_models(df, models, extra_model_args, parallel, plotose, **kwargs)
276 extra_model_args = extra_model_args or np.tile({}, len(models))
277 if parallel:
--> 278 traces = Parallel(n_jobs=min(os.cpu_count(),
279 len(models)))(delayed(split_train_predict)
280 (df, model, num_chains=1, **kwargs, **extra_model_arg)
~/env_jb1/lib/python3.8/site-packages/joblib/parallel.py in __call__(self, iterable)
1059
1060 with self._backend.retrieval_context():
-> 1061 self.retrieve()
1062 # Make sure that we get a last message telling us we are done
1063 elapsed_time = time.time() - self._start_time
~/env_jb1/lib/python3.8/site-packages/joblib/parallel.py in retrieve(self)
938 try:
939 if getattr(self._backend, 'supports_timeout', False):
--> 940 self._output.extend(job.get(timeout=self.timeout))
941 else:
942 self._output.extend(job.get())
~/env_jb1/lib/python3.8/site-packages/joblib/_parallel_backends.py in wrap_future_result(future, timeout)
540 AsyncResults.get from multiprocessing."""
541 try:
--> 542 return future.result(timeout=timeout)
543 except CfTimeoutError as e:
544 raise TimeoutError from e
/usr/lib/python3.8/concurrent/futures/_base.py in result(self, timeout)
442 raise CancelledError()
443 elif self._state == FINISHED:
--> 444 return self.__get_result()
445 else:
446 raise TimeoutError()
/usr/lib/python3.8/concurrent/futures/_base.py in __get_result(self)
387 if self._exception:
388 try:
--> 389 raise self._exception
390 finally:
391 # Break a reference cycle with the exception in self._exception
ValueError: The least populated class in y has only 1 member, which is too few. The minimum number of groups for any class cannot be less than 2.
It looks like using including intercept actually hurts leave-one-out posterior predictive. Actually, so does including floor in the model. To bring this home, let’s only use the models that did not have a warning above:
from bayes_window.model_comparison import compare_models
compare_models(df_clean,y=window1.y,parallel=True,
models = {
'full_normal': window1.model,
'no_condition_or_treatment': window1.model,
'no-treatment': window1.model,
'no_group': window1.model,
},
extra_model_args = [
{'treatment': window1.treatment, 'group': window1.group},
{'treatment': None, 'condition': None},
{'treatment': None, 'condition': window1.condition},
{'treatment': window1.treatment, 'group': None},
])
Keep in mind though that we had to remove some data that had too few labels in order to make LOO work.
References¶
Gelman, A., & Hill, J. (2006), Data Analysis Using Regression and Multilevel/Hierarchical Models (1st ed.), Cambridge University Press.
Gelman, A. (2006), Multilevel (Hierarchical) modeling: what it can and cannot do, Technometrics, 48(3), 432–435.
McElreath, R. (2020), Statistical Rethinking - A Bayesian Course with Examples in R and Stan (2nd ed.), CRC Press.